Of Liquid Images and Vital Flux
Every epidemic emplaces us in molecular visualizations where iconic spiked orbs circle host cells. As we become accustomed to SARS-CoV2, my essay returns to visualizations of the HIV-1 macromolecule to explore the role of dynamic images in scientific research. The fact that scientists are still building models of HIV-1, after four decades, should give us pause in our impatience about “knowing” SARS-CoV2 fully. The liquid image built on a synthetic-biologic continuum, I argue, provides a clue to the essentially speculative nature of visualizing and modeling virus-host vital relations. Malleable, dynamic, and always emergent, the liquid image races to visualize and predict the vital flux of parasitic relations, and to keep abreast of fast-paced research. I trace editable models and simulations of HIV through an analysis of cellPACK, a software suite developed at the Center for Computational Structural Biology at the University of California, San Diego. Integrating crowd-sourced data, the platform enables an image composed at one scale to continuously liquidate and reassemble into another (perhaps larger) whole, and vice versa. Such scalable images are creative actualizations of scientific hypotheses based on mathematical possibilities. And yet. Even as they render the capacities and tendencies of complex systems calculable, the partially known haunts the liquid image.
We encounter an ominous spiked orb almost on a daily basis in reportage on the COVID-19 pandemic. Those spikes attach with ease to the ACE2 receptors of human respiratory cells, we are told, enabling a deadly viral takeover of host resources. But we’ve heard this story before: SARS-CoV2 does to respiratory cells what HIV once did to CD4 cells of the immune system. An abiding accompaniment to every pandemic is the visualization of virus macromolecules, their approach and entry, bloom and blast in host cells. Despite the intense desire to separate and isolate an “invisible enemy,” we have come to regard pathogenic particles in their vital relation to the hosts that they occupy. Moving images based on structural data plunge us into wondrous voyages in which orbed bodies circle and encounter each other (Fig.1). As they morph and dissolve, we watch virus-host co-emergences in the twenty-first century “molecular fantastic.”[1] Operating on a synthetic-biologic continuum, these moving images are vital media, whose irreducible vitality is most evident in their liquid character. Plastic, dynamic, and speculative, the liquid image, I argue, races to visualize and predict the vital flux of parasitic relations.
We see this in the ongoing technical-aesthetic mediation of the HIV-1 macromolecule, still under construction after four decades of the viral pandemic. On the one hand, malleable, constantly editable images are necessary for processing volumes of new data in fast-paced scientific research. The “image” makes large volumes of data cohere as a single insight, a process that statistician and artist Edward Tufte (2001) named “de-quantification.”[2] Scientists can keep abreast of structural data from multiple sources in contributing to editable models of living systems. On the other, the speculative nature of the liquid image is strategic in keeping up with vital flux. One might consider how genetic mutations that change instructions for protein assembly will impact the entry of viral biomolecules into host cellular receptors—an understanding critical to drug design. For the “best understood of any organism,” as illustrator David Goodsell puts it, HIV clocks over 90 drug resistant genetic mutations.[3] Scientists continue to visualize and simulate HIV “molecular docking events” at host cellular sites that enable the design of new drugs—molecules which can sit in the docking site and block viral entry.[4] Dynamic speculative images keep pace with evolving virus-human emergences in these molecular-cellular contexts.
[Fig.1] Screenshot from “HIV Life Cycle” [5]
The mediatic capture of HIV-1 emerges at the intersection of basic science laboratories and creative media industries. While media histories starting from the first capture of the tobacco mosaic virus under the electron microscope in 1938 reveal a shift from morphological preoccupations to decoding viral genomes in the mid-twentieth century, biomolecular models of virus macromolecules remain critical to integrative research.[6] No doubt the push arrives with the turn to complex systems thinking in the life sciences that made computation central to the study of living organisms. In attempting to understand the effects of vital processes at one scale (e.g. biochemical cellular reactions) on another (e.g. folding of protein molecules), scientists of different cloth increasingly share structural data and test competing hypotheses, communicate and educate on new media platforms such as (Maxon’s) Cinema 4D or (Autodesk’s) Maya. The “molecular movies,” as they are dubbed, are robust collaborations between academic researchers, biotech corporations, and digital animators.[7] Making the unseen visible, as the SciVis mantra goes, moving images integrate structural data and simulate possible molecular events in this post-cinematic context. With new data inputs come new possibilities.
In liquid images, “life” as vital process appears as “life itself.” As shorthand, “life itself” refers to the mediatic appearance of vital processes.[8] Life “emerges with” matter: bodily media like blood or respiratory mucosa sustain the virus. Technical-aesthetic mediation detects and composes the interactions of viral particles in bodily fluids as the numeric distribution of x copies in y milliliter blood (à la the viral load test). Sarah Kember and Johanna Zylinska (2010) name this mediatic capture a “cut” in the flow of living processes—a slice, a snapshot—so that “life” makes an appearance as “life itself.” In liquid images, life appears vibrating, morphing, changing. Animations of dynamic processes make vital flux legible and palpable in multisensory environments.[9]
The point is better made with brief illustration: I focus on the making of HIV macromolecules at the Center for Computational Structural Biology (formerly, Molecular Graphics Lab) at the Scripps Research Institute, University of California, San Diego. Its chief biologist, Arthur Olson, was a pioneer in the computer graphics foundational to molecular visualizations. Founded in 1981, the “Olson Lab” (as it is nicknamed) sources its data from molecular biologists (studying gene transcriptions), biochemists (studying chemical interactions), and structural biologists (studying protein assemblies) to build integrative crowd-sourced models which allow researchers to move beyond their silos of expertise.[10] Until now, the lab has made six models of the HIV-1 macromolecule on media platforms so that researchers can manually move molecules around, watch for folds, accelerate or decelerate motion. While the game industry’s VR platforms provide inspiration for analyzing biomolecular structures in 3-D, sophisticated toolkits specifically tailored for biological research are now available; some of these have been developed at the Olson Lab.[11] A biology-specific extension of the better-known AutoPACK is a suite of programs called cellPACK, which was developed in collaboration with medical-illustrator-turned-software-designer Graham Johnson (Mesoscope Lab, University of California, San Francisco).[12] Structural biologist and illustrator David Goodsell’s legendary “zoomable” watercolors, which magnified cellular structures into their molecular components, are the methodological and aesthetic bases for building unified hybrid models on cellPACK (Fig.2a).[13]

[Fig.2a] Watercolor of HIV in Blood Plasma, 1999
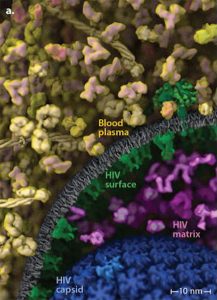
[Fig.2b] HIV-in-Blood Plasma Visualization, 2015
The idea is to combine data from all branches of biology into a comprehensive mesoscale model ranging from 0.1 micron to 10 nanometers. “Artistic depictions of cellular environments” built on the cellPACK suite can integrate data from ultrastructural (light and electron microscopy), infrastructural atomic (x-ray crystallography and NMR spectroscopy), and biochemical (for concentrations and interactions) sources (Johnson et al. 2015, 85-91). “Ingredients” from these data sources produce scientific “recipes” (as the lab’s extended cooking metaphor runs) for various parts of the model. Fig.2b is an image, HIV-in-Blood Plasma, made up of seven different recipes (each with a different color coding) that are unified into a single model. Endless updates to the “editable model of HIV” become possible in these plastic, malleable liquid images. The Olson Lab’s HIV-1 ultrastructure (the HIV envelope) functions as the polyhedral “mesh” or “state space” (adding time to geometric coordinates) for packing ingredients (De Landa 2011). This is accomplished by cellPACK’s virtuoso “packing algorithms” that determine how the mesh is filled out to produce a scalable image (see Fig.3).
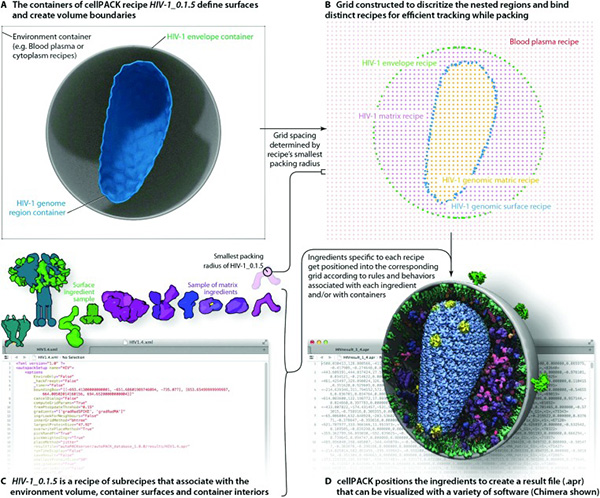
[Fig. 3] Packing the HIV-1 mesh
Such a drive toward consistent precision in modelling biomolecular structures enables the study of “active sites” on cellular surfaces that drug therapies target to block the successful “docking” of HIV. Stefano Forli, who is the biotech interface at the lab, explained how animating 3-D models on new media platforms not only provided the opportunity to circle, view, and analyze cellular structures, but it also enabled calculating the densities, forces, and propensities of molecules as they interacted with each other. Real-time video-tracking allowed for moving molecules around, accelerating or decelerating molecular motion. As the molecules vibrated in Brownian motion, Olson noted, scientists could better understand the electrostatic complementarity and hydrogen bonding that forms molecular structures. They could zoom into specific sites of structural uncertainty and then out to the bigger picture—the HIV-1 viral capsid in the extracellular environment.
This example of liquid images discloses their infinite malleability. To be sure, these “technical images,” to recall Vilém Flusser (1985), concretizing a “universe of particles moving toward dissipation,” are as plastic as are all digital images. Each image liquidates and reconfigures the previous trace. And yet, the point of the constant update is to keep up with high volumes of incoming data: the Olson Lab’s release of HIV 1.6 demonstrated the possibility of integrating crowd-sourced structural data at speeds commensurate with the fast pace of research (Johnson et al. 2014, 23-44). Further, when animated, scientists were able to experiment with multiple outcomes for the molecules in motion—colliding, adhering, separating. Such dynamism is critical to simulating the changeful interactivity of parasitic relations. As obligate parasites, bits of nucleic acid with a protein coat and without cell walls, we know that viruses “come alive” as secondary organisms always in vital relation to a host; they need host resources to replicate.[14] But this biological partnership is constantly negotiated as virus-host vital relations continue to emerge.[15] Computer animations can simulate the processualities of emergences, predicting a range of probable outcomes for the form and evolution of vital relations.
Investigating simulations of living systems, Manuel De Landa (2011) examines the computational rendering of biologic “possibility spaces”—the space of genes, the space of structural proteins, the space of spatial structures—into synthetic possibility spaces. The efficacy of the scalable liquid image lies in rendering biological possibilities as calculable mathematical probabilities. In this enriched mechanistic account of life, De Landa asks: What “individual singularities” arise in interactions between agents within a possibility space? Defining individual singularities as new properties, De Landa argues that hydrogen and oxygen produce a liquid state more than the sum of its parts, a state whose properties (such as the temperature at which water boils) are distinct from the two gaseous entities. Secondary mutations of HIV that evolve as drug resistance are examples of such individual singularities: these properties emerge at negotiations between host resources, viral particles, and chemical drug molecules. While such new properties can be decoded and documented, what remains speculative are the capacities and tendencies that come into play within living systems. Enter liquid images: the attempt to predict outcomes of new individual singularities arising in cellular environments.
As an image composed at one scale liquidates to reassemble into another, perhaps larger, whole, or vice versa, liquid images not only present variable trajectories but they also predict what is not yet known. They are crucial sites for creative actualizations of scientific hypotheses based on mathematical possibilities. Whether those hypotheses concern envisioning the capacities and tendencies of complex systems, or tracking evolving changes in new singularities, the partially known haunts the liquid image. That said, decades of research on HIV forms a baseline for imaging SARS-CoV2. We are indeed ahead of the game, even as it may not seem like it. The famous David Ho, innovator of the protease inhibitors of the anti-retroviral therapies, has been tapped for Joe Biden’s team on COVID-19 for a reason. But to expect definitive findings in a matter of months is to live in a fool’s paradise. We know scientific agreements on particular outcomes proposed in scientific visualizations come after years of study. Even though HIV is relatively simple in its composition (made of just eight structural proteins), the “HIV trimer” was on the “most wanted” list of protein macromolecules until 2013, when the scientific community could agree on its definitive crystalline structure. The liquid image is an object lesson in patience.
We will have to inhabit the molecular fantastic of SARS-Cov2 for the foreseeable future, watching as this increasingly familiar orb circles and enters its new hosts, leaving cellular ruin in its wake. The liquid image is always at a lag for vital flux continues to exceed all synthetic transcription. Delving deeper, moving faster, these speculative images remain in hot pursuit of ever-emergent virus-host vital relations.
References
Beckman, Karen, ed. 2014. Animating Film Theory. Duke University Press.
De Landa, Manuel. 2011. Philosophy and Simulation: The Emergence of Synthetic Reason. New York, London: Continuum.
Douglas, Angela. 1987. The Biology of Symbiosis. Princeton University Press.
———.1994. Symbiotic Interaction. Oxford University Press.
———.2010. The Symbiotic Habit. Princeton University Press.
Ghosh, Bishnupriya. 2016 “Toward Symbiosis: Human-viral Futures in the “Molecular Movies,” In Sustainable Media: Critical Approaches to Media and Environment, edited by Nicole Starosielski and Janet Walker, 232-247. Duke University Press 2016.
Johnson, Graham T. et al. 2014. “3D molecular models of whole HIV-1 virions generated with cellPACK,” Faraday Discuss, Nov 23: 23–44.
Johnson, Graham T. et al. 2015. “cellPACK: A virtual Mesoscape to model and visualize structural systems biology,” Nature Methods, 12.1: 85-91.
Kember, Sarah, and Johanna Zylinska. 2010. Life after New Media: Mediation as Vital Process. MIT Press.
Myers, Natasha. 2015. Rendering Life Molecular: Models, Modelers, and Excitable Matter. Duke University Press.
Pickford, Clifford, and Stuart K. Tewksbury. 1994. Frontiers of Scientific Visualization. Wiley-Interscience Pub.
Tufte, Edward. 2001. The Visual Display of Information. Graphics Press.
Notes
[1] With advanced imaging technologies, in scientific edutainment, we are in the domain of the marvelous: a molecular fantastic of the same order as Akira Lippit’s “optical fantastic” in the first half of the 20th (triangulating the development of X-ray technologies, the splitting of the atom, and psychoanalysis, see Atomic Light (Shadow Optics), Minnesota University Press, 2005). See my account of the cellular agon in “Animating Uncommon Life: U.S. Military Malaria Films (1942-1945) in the Pacific Theater,” in Beckman 2014.
[2] Edward Tufte, The Visual Display of Information (Graphics Press, 2001).
[3] Stanford University HIV Drug Resistance Database (https://hivdb.stanford.edu/).
[4] Scientific visualizations that model crowd-sourced data play a critical role in antiviral strategies of epidemic mediation. The histories of SciVis are vast because they include material practices obtaining across computer graphics, bioscience laboratory techniques, and virtual art. I discuss their conjugation further in my current book project, The Virus Touch: Theorizing Epidemic Mediation.
[5] For the animation, see https://vimeo.com/260291601. This was made collaboratively with the Department of Biochemistry at the University at Utah and the CHEETAH consortium (https://biochem.web.utah.edu/iwasa/projects/HIV.html). Janet Iwasa (Animation Lab), whose scientific animations I have explored elsewhere, headed the project with data sourced from researchers globally. See, Ghosh 2016.
[6] 100-500 times smaller than bacteria, viruses were the microbes that passed through Louis Pasteur’s porcelain filters and remained unobservable under ordinary light microscopes. When the powerful electron microscope made optical capture possible, research on identifying viruses began in earnest. The focus on virus morphologies soon gave way to the romance with genomic codes in the mid-20th century as molecular biologists drilled down to genomic fingerprints of viruses in host DNA. The latter remains the bedrock of virological research, even as, still later in the 20th century, with the rise of systems biology, the onus fell on integrating research.
[7] The mantra is associated with SciVis practices which begins to commercially cohere in the late eighties (the precedents in molecular graphics go back much further). The first SciVis congregation (the Visualization in Scientific Computing Conference), in 1987, brought together industry, academics, and government officials. Thus, began the industrial enterprise of new media platforms that allowed scientists and animators to produce images based on experimental data necessary for designing source materials (digital files for synthetic compounds) that could be actualized as marketable “biologicals” (biotechnological products). See, introduction to Pickford and Tewksbury 1994.
[8] We might remember that “emergence,” which is a key concept for large-scale ecological disturbances such as new pathogenic species relations, comes from the Latin root emegere signifying “to appear.” In this regard, studying viruses in an agar plate under the electron microscope is an epistemic cut that makes virus-human relations legible, readying them for other interventions such as gene-splicing or drug therapy.
[9] For the multisensory dimension of building protein macromolecules, one of the best studies is Natasha Myers’ (2015) ethnography of protein crystallography. Myers traces the haptic and kinesthetic qualities of modeling proteins to argue for a sensuous engagement with scientific molecular structures.
[10] Arthur Olson has been envisioning HIV from the start of the epidemic in the United States. Studying the structural dimensions of HIV, the Olson Lab’s “AIDS-Related Structural Biology Program” is an NIH-funded operation. The team at the lab includes a range of scientists from David Goodsell (on the technical-aesthetic end) to Stefano Forli (on the biotech industry end). The lab developed at least one thousand structures of protease, one of the three important HIV enzymes in the viral replication cycle. Research on HIV enzyme biostructures has everything to do with the life-saving protease inhibitors, placing the Olson Lab squarely within ongoing efforts to develop pharmacological solutions to “managed HIV” (as it is now called).
[11] In the area of software programs, the lab’s successes are many. “Autodock,” a program for biomolecular structure analysis, has been adopted by four thousand laboratories all over the world.
[12] cellPACK was developed at the Olson Lab (with Ludovic Antin) while Johnson was finishing his post-graduate work. The open-source program integrates structural biology and systems biology data with packing algorithms to assemble comprehensive 3D models of cell-scale structures in molecular detail.
[13] David Goodsell is a central figure who is well known for his watercolor illustrations based on real data sources and magnification specifications; these are available on the Protein Data Bank. Goodsell has not only led on the scientific front, but his books, The Machinery of Life (1993) and Atomic Evidence (2016), are widely considered exemplars of science communication. Based on his aesthetic protocols, the Olson Lab has produced as many as 6 HIV-1 models to date (HIV-1 1.2, 1.3…), each manually curated with widely sourced data.
[14] Ever since its discovery in the late 19th century, the virus has always been a border object, in the sense that it lacks an important definitive feature of living organisms—the capacity to metabolize what it needs to regenerate. One of the most influential definitions of “life” hearkens to physicist Erwin Schrödinger’s simple but lasting distinction of living things as irritable, and capable of reproduction and metabolism. As opposed to crystals, living things were defined, Schrödinger maintained, by their capacity for self-regeneration (to grow, repair, and reproduce) and their fight against dissipative entropy (“What if Life?” 1941). An obligate parasite is an organism that cannot live without a host, as opposed to a facultative parasite that can live independently but becomes parasite under certain conditions. The virus can only live within hosts. It literally “comes alive” from dormant crystalline states when it senses a host.
[15] One of the foremost scholars of parasitism, Angela Douglas characterizes symbiosis as a derived state: a gradual evolutionary transition from antagonism (including virulent pathogenesis) to mutually beneficial relations including a stable, managed partitioning of resources. One partner, usually the host, takes control of resource distribution over time, imposing sanctions and controlling transmissions for both partners, even as both organisms develop novel capacities (a lateral, not hereditary, transfer of properties) in order to live with the perturbations that the other generates. Symbiosis-at-risk is one step on a spectrum of relations between parasites and hosts; and some relationships (e.g. human-Ebola) never move past that point. Symbiotic relations are those that are mutually beneficial to the participants for the major duration of their lifetime. This does not mean that parasitism is not symbiotic, but that pathogenic parasitism is not. Less virulent parasites are at a selective advantage in this regard, since they do not deplete the resources of the host. We can “live with” virulent pathogens like HIV only with the technological mediation of drug therapies. See, Angela Douglas, The Symbiotic Habit (Princeton UP, 2010), Symbiotic Interaction (1994), and The Biology of Symbiosis (1987).